OpenVax
Open source software for personalized cancer vaccines
The OpenVax group develops open source software for designing personalized cancer vaccines. Our work includes a bioinformatics pipeline for selecting a peptide vaccine from DNA and RNA sequencing of a patient's tumor, as well as a collection of Python libraries developed to support it (see our GitHub page). We have helped initiate and run several clinical trials of personalized cancer vaccines at the Mount Sinai Hospital, in collaboration with Dr. Nina Bhardwaj, Dr. Adilia Hormigo, Dr. Matthew Galsky, and Mount Sinai's Vaccine and Cell Therapy Lab.
Contributors



Publications

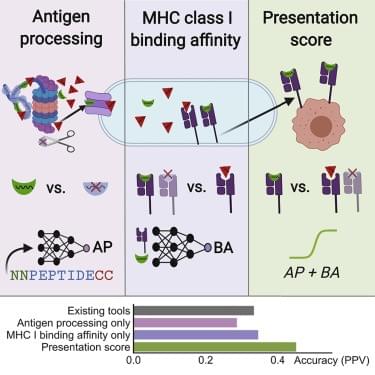
We developed an integrated predictor of MHC class I presentation that combines new models for MHC class I binding and antigen processing. Considering only peptides first predicted by the binding model to bind strongly to MHC, the antigen processing model is trained to discriminate published mass spectrometry-identified MHC class I ligands from unobserved peptides. The integrated model outperformed the two individual components as well as NetMHCpan 4.0 and MixMHCpred 2.0.2 on held-out mass spectrometry experiments. Our predictors are implemented in the open source MHCflurry package, version 2.0 (github.com/openvax/mhcflurry).

This paper describes the sequencing protocol and computational pipeline for the PGV-001 personalized vaccine trial. PGV-001 is a therapeutic peptide vaccine targeting neoantigens identified from patient tumor samples. Peptides are selected by a computational pipeline which identifies mutations from tumor/normal exome sequencing and ranks mutant sequences by a combination of predicted Class I MHC affinity and abundance estimated from tumor RNA. The PGV pipeline is modular and consists of independently usable tools and software libraries. We hope that the functionality of these tools may extend beyond the specifics of the PGV-001 trial and enable other research groups in their own neoantigen investigations.

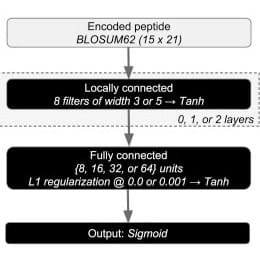
Predicting the binding affinity of major histocompatibility complex I (MHC I) proteins and their peptide ligands is important for vaccine design. We introduce an open-source package for MHC I binding prediction, MHCflurry. The software implements allele-specific neural networks that use a novel architecture and peptide encoding scheme. When trained on affinity measurements, MHCflurry outperformed the standard predictors NetMHC 4.0 and NetMHCpan 3.0 overall and particularly on non-9-mer peptides in a benchmark of ligands identified by mass spectrometry. The released predictor, MHCflurry 1.2.0, uses mass spectrometry datasets for model selection and showed competitive accuracy with standard tools, including the recently released NetMHCpan 4.0, on a small benchmark of affinity measurements. MHCflurry's prediction speed exceeded 7,000 predictions per second, 396 times faster than NetMHCpan 4.0. MHCflurry is freely available to use, retrain, or extend, includes Python library and command line interfaces, may be installed using package managers, and applies software development best practices.
Clinical Trials
Personalized cancer vaccine trials at Mount Sinai using our software
Principal Investigator: Nina Bhardwaj
Safety & immunogenicity trial for the PGV-001 vaccine, which consists of 10 neoantigenic peptides injected with an adjuvant (poly-ICLC). Broad set of malignancies: H&N, NSCLC, breast, ovarian, bladder, SCC, multiple myeloma. Patients must be disease-free when beginning treatment with PGV-001, so this trial is most applicable following a complete resection.
Principal Investigator: Adilia Hormigo
The PGV-001 neoantigen peptide vaccine in combination with standard-of-care temozolomide and Novocure's "tumor treating fields" for glioblastoma.
Principal Investigator: Matt Galsky
The PGV-001 neoantigen peptide vaccine in combination with an anti-PD-L1 checkpoint agent (atezolizumab) for bladder cancer.
Open Source Software
This is the public version of the OpenVax bioinformatics pipeline for selecting patient-specific cancer neoantigen vaccines, which is currently the basis for the three personalized vaccine clinical trials listed above.
Vaxrank is used to generate a ranked list of personalized cancer vaccine peptides from somatic mutations and tumor RNA sequencing data. The most distinctive feature relative to other similar tools (MuPeXi, pVacSeq) is phasing of adjacent somatic/germline variants using a mutant coding sequence assembled from RNA reads.
Topiary is an older tool for predicting mutated peptide MHC ligands from somatic variants without RNA evidence.
MHCflurry predicts the antigens available for recognition by CD8+ T cells. The most recent version provides a pan-allele MHC I binding predictor as well as an experimental antigen processing predictor.
Isovar determines mutant protein sequences by assembling RNA reads which overlap and support somatic variants. Useful for correctly incorporating other adjacent variants (phasing) as well as mutation associated splicing differences (intron retention).
Varcode predicts mutant protein sequences from DNA variants with a friendly Python API.
Blog
Thoughts and research snippets
21 de outubro de 2020We've previously written about the OpenVax neoantigen prediction pipeline, which is the...3 de agosto de 2018Cancer immunotherapy aims to recruit the body's own immune system to attack a tumor, which...27 de junho de 2018 · immunologyUri Laserson organized a delightful class at Mount Sinai called Applications of DNA Sequencing...